I hold a Doctor of Philosophy degree from the Catholic University of America, with a specialization in the structural analysis of portal protein gp20. My research within the realm of T4 virus has been published in esteemed scientific journals and has also been featured on their covers. In the future, my academic pursuits will focus on investigating the molecular structures of bacteriophages, infection processes, and cellular structures.


Victor Padilla-Sanchez, PhD
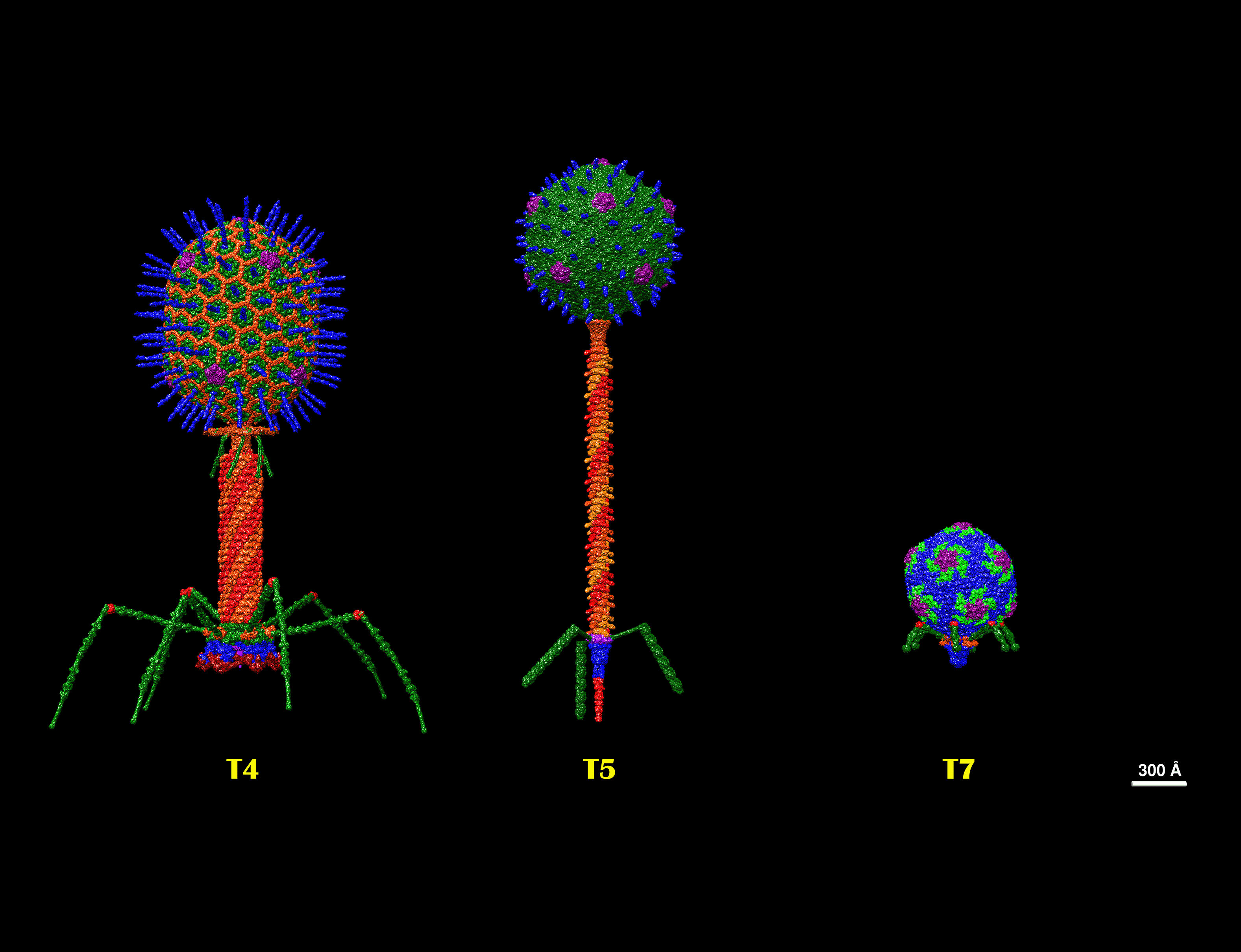
Victor Padilla-Sanchez, PhD
Order Caudovirales. Structures of T Bacteriophages representing the seven T types of Escherichia coli phages described by Max Delbruck in the 1940’s. T4 of the Myoviridae family, T5 of the Siphoviridae family, and T7 of the Podoviridae family. The structures were built from individual protein data bank (pdb) files in the UCSF Chimera software, which were updated to the year 2024 and at real scale.

Victor Padilla-Sanchez, PhD
COVID-19 infection is attributed to the SARS-CoV-2 virus, which belongs to the coronaviridae family. The atomic resolution structures of these viruses, as they interact with cell membrane and ACE2 receptors, have been studied. The outbreak of COVID-19 in 2019 and its subsequent pandemic have been linked to these coronaviruses, resulting in a significant number of deaths. Further information on this topic can be found in the following reference: Padilla-Sanchez V (2021) SARS-CoV-2 Structural Analysis of Receptor Binding Domain New Variants from United Kingdom and South Africa. Research Ideas and Outcomes 7: e62936.
Victor Padilla-Sanchez, PhD
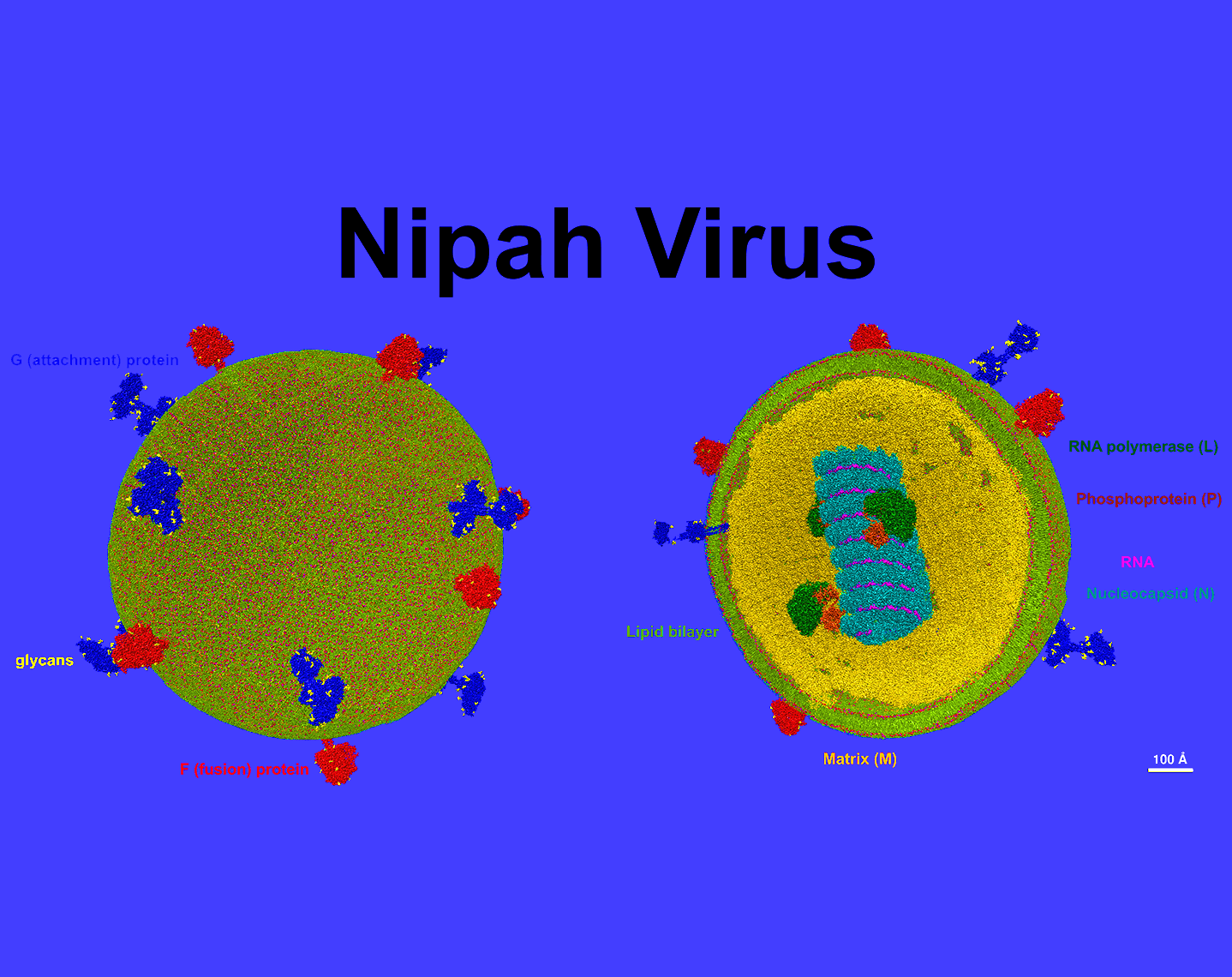
The Nipah virus structural model, constructed at an atomic resolution, depicts a particle with a diameter of 90 nm, adorned with spikes. This model affords a glimpse into the virus's interior. The Nipah virus is known for its high mortality rate and is viewed as a potential candidate for the next pandemic. The construction of this model utilized components from the UCSF Chimera database, sourced from the Protein Data Bank (pdb).
Victor Padilla-Sanchez, PhD
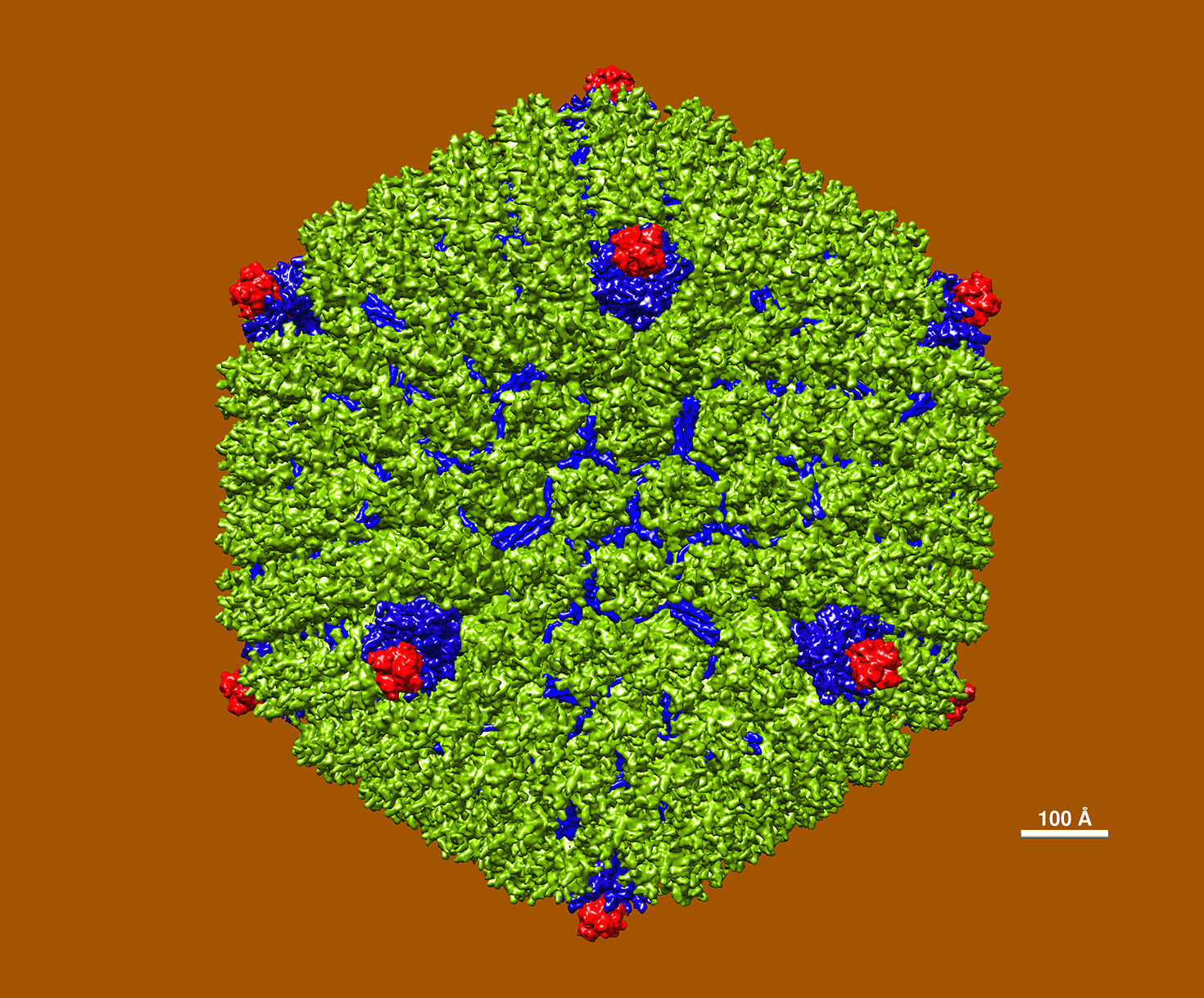
Adenoviruses belong to the family Adenoviridae and are characterized by their non-enveloped structure, which measures 90-100 nanometers in diameter, and has an icosahedral nucleocapsid containing double-stranded DNA. They were first identified in human adenoids in 1953, and their name is derived from this source. The image provided above shows the atomic resolution model of Adenovirus B35. These viruses are known to target the respiratory tract, conjunctiva, and urinary tract.
| The central objective of my research is the structural analysis and molecular dynamics of T Bacteriophages that infect Escherichia coli. Furthermore, I am investigating HIV-1 vaccine development and analyzing the molecular interactions of SARS CoV-2, herpes virus, and various bacteriophages such as SPP1, phi29, and lambda, as well as mycobacteriophages. A crucial component of my research is the application of software such as UCSF Chimera and VMD/NAMD for structural and macromolecular complex analysis at atomic resolution, including viral infections and entire organisms. My work has been extensively featured in scientific publications and news outlets worldwide over the last two decades, indicating its significance to both scientific and medical communities. |
-
Research
-
Publications
-
Courses
-
CATCMB
-
Gallery
-
Lab Members
-
Organization
-
Links